Understanding transport through ion channels by molecular dynamics simulations and thermodynamic integration
With the crystal structures of membrane proteins becoming available and an increasing computer power, classical molecular dynamics simulations have contributed to a qualitative understanding and to the quantitative computation of transfer rates of ion channels. With typical transfer rates of the order of 106 to 108 s-1, however, the number of events is too small to form the basis of a proper statistical analysis or to observe any event at all. Hence, the computation of potentials of mean force (or free energy landscapes) as a function of a small set of effective coordinates has become the standard tool to determine an approximate free energy surface for ion transfer. Using the characteristic free energies of the transfer process, rate equations can be formulated to estimate the overall turnover.
Based on our experience on electron transfer in membrane proteins[1-3], we advance the method of thermodynamic integration (TI) and apply it to ion-conducting channels of increasing complexity. TI calculations aim at computing the difference in free energies between two states A and B, here preferential sites of ion localization. Additional to the two potential energies UA and UB, a coordinate Λ is used, which couples the initial and product states so that the transition from A to B can be calculated along this non-physical reaction coordinate. A mixed potential U(Λ) results, chosen so that it corresponds to UA for Λ=0 and UB for Λ=1, U(Λ)=f(Λ)UA+(1-f(Λ))UB. In this scheme, the free energy difference between the two states can be described as:
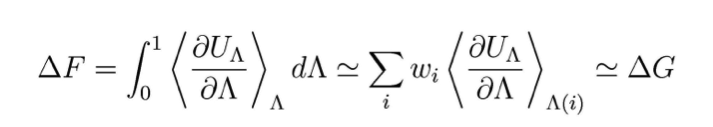
Boltzmann-weighted averages of this function are generated by molecular dynamics simulations for a finite number of points along the curve. Our calculations are based on the Amber force field and program package, and we include water and a representative lipid bilayer in the simulations.
We investigate prototypical gramicidin A double helical dimer in its two polymorphs, as there exists a broad spectrum of experimental data and computer simulations. With four co-crystallized Cs+ ions, we have a sound structural basis for the intended thermodynamic integration calculations. Subsequently, we wish to turn to ammonium transporters. Finally, we plan to have a look at proton transfer in complex I. Here, our objective is to compute the free energy landscape for proton transport in the two channels closest to the quinone binding site.
References
[1] C. Wittekindt, M. Schwarz, T. Friedrich, T. Koslowski (2009) JACS 131, 8134.
[2] F. Burggraf, T. Koslowski (2014) BBA Bioenergetics 1837, 186.
[3] A. Bauß, T. Koslowski (2015) PCCP 17, 4483.
Publications
Na, S, Bauß, A, Langenmaier, M, Koslowski, T (2017) Thermodynamic Integration Network Study of Electron Transfer: from Proteins to Aggregates, PCCP, 19, 18938.
Contact
Prof. Dr. Thorsten Koslowski
Institute of Physical Chemistry
Albertstr. 23a
79104 Freiburg
Phone: +49 (761) 203 6182
Fax: +49 (761) 203 6189
Thorsten.Koslowski@physchem.uni-freiburg.de
http://www.theochem.uni-freiburg.de/



