Proton translocation by respiratory complex I
The proton-pumping NADH:ubiquinone oxidoreductase, respiratory complex I, links the electron transfer from NADH to ubiquinone with the translocation of protons across the membrane. In doing so, complex I establishes a proton motive force required for energy consuming processes. In bacteria, the complex consists of 14 different subunits named NuoA through NuoN. The structure of the complex from several organisms was resolved by x-ray crystallography and cryo-electron microscopy at molecular resolution [1,2]. A peripheral arm containing all cofactors for the electron transfer reaction protrudes into the aqueous phase and a membrane arm catalyzing proton translocation is buried in the lipid bilayer.
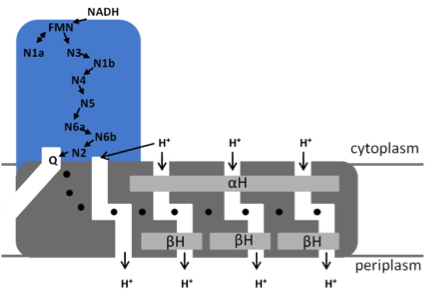
Model of the bacterial NADH:ubiquinone oxidoreductase (complex I) N1a to N7 represent the Fe/S clusters, Q, ubiquinone. The position of the quinone-binding chamber, the four proton pathways, the central hydrophilic axis (•) and the stabilizing elements (αH and ßH) are indicated.
NADH is oxidized by FMN at the tip of the peripheral arm. The electrons are transferred via a series of 7 iron-sulfur (Fe/S) clusters to the quinone-binding site located at the interface of the two arms [3]. Cluster N1a is not part of that „electron wire‟ to the quinone-binding site but regulates NADH oxidation by temporarily accepting one electron from the FMN [4]. The membrane arm contains a unique 110 Å long horizontal helix (αH) aligning to the membrane arm that acts as a clamp to hold the subunits catalyzing proton translocation together [4]. Four proton channels connected by a central axis of charged amino acids have been detected in the membrane arm [5]. The coupling between the spatially separated electron transfer and proton translocation is not understood, although it is discussed that the redox chemistry of the most distal Fe/S cluster N2 and the quinone drive proton translocation.
The aim of this project is to gain insight into the structure and function of complex I, especially concerning the coupling of redox chemistry with proton translocation, through a combination of genetic, biochemical and biophysical approaches. Specifically, we will elaborate the role of the distal Fe/S cluster N2 for proton translocation and identify amino acid residues and structural elements in the quinone-binding cavity essential for proton translocation.
The most distal Fe/S cluster N2 is distinguished from all other clusters of the complex by its more positive and pH dependent midpoint potential and by its unusual ligation by two vicinal cysteine residues. It was proposed that this motive is essential for proton translocation. We will change the binding motif of N2 to a regular motive for binding a tetranuclear Fe/S cluster by site-directed mutagenesis and determine the electron transfer and proton translocation activity of the mutants. The spectroscopic properties of the cluster will be determined by helium-temperature EPR-spectroscopy.
The quinone-binding chamber of complex I has a length of about 30 Å and the quinone headgroup is located approximately 15 Å from the membrane surface. A narrow apparent entrance leads to the chamber that is completely enclosed from the solvent [1]. The chamber is faced with patches of hydrophobic and hydrophilic amino acid residues, respectively. The ionizable groups within the chamber are highly conserved and mutations in some of them led to inactive variants. In addition, the quinone-binding chamber is connected to the central hydrophilic axis of the membrane that connects the proton channels by a chain of charged residues. In order to determine whether mutations in the binding-chamber or in the chain of charged residues have an influence on proton translocation, we will systematically exchange the ionizable amino acids in the chamber and the chain of charged residues and select for those mutations that are capable of electron transport. The complex will be isolated from these strains and characterized by enzyme kinetics and EPR spectroscopy. The electron transfer and proton translocation rate of the preparations will be determined.
References
[1] Baradaran R, Berrisford JM, Minhas GS, Sazanov LA (2013) Nature494:441-445.
[2] Agip A-N, Blaza JN, Fedor JG, Hirst J (2019) Annu. Rev. Biophys. 48:165-184.
[3] De Vries S, Dörner K, Strampraad MFJ, Friedrich T (2015) Angew. Chem. Int. Ed., 54:2844-2848.
[4] Schulte M, Frick K, Gnandt E, Jurkovic S, Burschel S, Labatzke R, Aierstock K, Fiegen D, Wohlwend D, Gerhardt S, Einsle O, Friedrich T (2020) Nature Commun.10:2551.
[5] Steimle S, Schnick C, Burger EM, Nuber F, Krämer D, Dawitz H, Brander S, Matlosz B, Schäfer J, Maurer K, Glessner U, Friedrich T (2015) Mol. Microbiol., 98, 151-161.
[6] Mühlbauer M, Saura P, Nuber F, Di Luca A, Friedrich T, Kaila V (2020) J. Am. Chem. Soc.accepted, DOI: 10.1021/jacs.0c02789.
Publications
-
Dörner K, Vranas M, Schimpf J, Straub IR, Hoeser J, Friedrich T (2017) Significance of the [2Fe-2S] Cluster N1a for Electron Transfer and Assembly of Escherichia coliRespiratory Complex I. Biochemistry56:2770-2778. DOI: 10.1021/acs.biochem.6b01058.
-
Gnandt E, Schimpf J, Harter C, Hoeser J, Friedrich T (2017) Reduction of the off- pathway iron-sulphur cluster N1a of Escherichia coli respiratory complex I restrains NAD+ dissociation. Sci. Rep.7:8754. DOI:10.1038/s41598-017-09345-4.
-
Na S, Jurkovic S, Friedrich T, Koslowski T (2018) Charge transfer through a fragment of the respiratory complex I and its regulation: an atomistic simulation approach. Phys. Chem. Chem. Phys.20:20023-20032. DOI: 10.1039/c8cp02420k.
-
Schulte M, Frick K, Gnandt E, Jurkovic S, Burschel S, Labatzke R, Aierstock K, Fiegen D, Wohlwend D, Gerhardt S, Einsle O, Friedrich T (2020) ) A mechanism to prevent production of reactive oxygen species by Escherichia colirespiratory complex I. Nature Commun.10:2551.DOI: 10.1038/s41467-019-10429.
-
Santos Seica AF, Schimpf J, Friedrich T, Hellwig P (2020) Visualizing the movement of the amphipathic helix in the respiratory complex I using a nitrile infrared probe and SEIRAS. FEBS Lett.594:491-496. DOI: 10.1002/1873-3468.13620.
-
Sonn-Segev A, Belacic K, Bodrug T, Young G, VanderLinden RT, Schulman BA, Schimpf J, Friedrich T, Dip PV, Schwartz TU, Bauer B, Peters J-M, Struwe WB, Benesch JLP, Brown NG, Haselbach D, Kukura P. (2020) Quantifying the heterogeneity of macromolecular machines by mass photometry. Nature Commun.11:1772. DOI: 10.1038/s41467-020-15642-w.
Contact
Prof. Dr. Thorsten Friedrich
Institute of Biochemistry
Albertstr. 21
79104 Freiburg
Phone: +49 (761) 203 6060
Fax: +49 (761) 203 6096
friedrich@bio.chemie.uni-freiburg.de
http://www.bioenergetics.uni-freiburg.de



