Method courses
Analysis of protein transport across mitochondrial membranes
We will generate radiolabelled precursor proteins by in vitro translation and import them into isolated mitochondria. Various analytical tools will then allow us to follow their import and intraorganellar sorting to the respective subcompartiment.
|
Prerequisites: |
|
Basic knowledge of RNA- and protein biochemistry. | |
|
Teaching goals: |
|
Understanding principles of posttranslational protein import and sorting. | |
| Instructor: | Prof. Dr. Chris Meisinger | ||
| Duration: | 2-3 days | ||
| Participants: | 3-4 | ||
| Location: | Institute of Biochemistry and Molecular Biology |
Biophysical characterization of protein stability and molecular interactions by Differential Scanning Fluorimetry (nanoDSF)
NanoDSF is a rapid and precise tool to analyze protein stability and molecular interactions. It requires little amount of protein and can be used for soluble and membrane proteins, for instance for optimization of protein purification protocols, detergent screening or ligand binding analysis. Protein samples can be brought for analysis.
|
Prerequisite: |
Basic knowledge of protein biochemistry. |
|
Teaching goals: |
Understanding principles and applications of nanoDSF |
|
Instructor: |
Dr. Evgeny Mymrikov |
|
Duration: |
1 day |
|
Participants: |
3 students |
|
Location: |
Institute for Biochemistry and Molecular Biology (AG Hunte) |
Biophysical characterization of molecular interactions by Microscale Thermophoresis (MST)
MST is a biophysical analysis tool which can be used to characterize biomolecular interactions of proteins, especially protein-protein interactions, in solution. Benefits are the usability for a broad range of binding affinities, little protein consumption, good applicability for membrane proteins, and typically rapid assay development time. Protein samples can be brought for analysis.
|
Prerequisites: |
|
Basic knowledge of protein biochemistry. | |
|
Teaching goals: |
|
Understanding principles and applications of MST. | |
| Instructor: | Simon Fuchs | ||
| Duration: | 1 day | ||
| Participants: | 3 students | ||
| Location: |
Institute of Biochemistry and Molecular Biology (AG Hunte) |
Blue native gel electrophoresis
Blue native gel electrophoresis is a powerful tool to separate and characterize protein complexes of biological samples like mitochondria. This technique allows the analysis of the stability, activity and assembly of protein complexes.
| Prerequisites: | Basic knowledge of gel electrophoresis like SDS-PAGE and Western blotting. | ||
| Teaching goals: |
Understanding principles and applications of blue native gel electrophoresis. | ||
| Instructor: | Dr. Sabrina Oppermann | ||
| Duration: | 2 days | ||
| Participants: | 3-4 students | ||
| Location: | Institute for Biochemistry (AG Friedrich) |
Cell-free production of membrane proteins
In contrast to soluble proteins membrane proteins tend to aggregate upon their translation in typical conventional expression systems. We will learn in this course how to overcome this problem using various cell-free translation systems deriving from rabbit reticulocyte lysates or wheat germ lysates. The membrane proteins can be kept in theses systems in competent forms e.g. for insertion into biological membranes or artifical liposomes.
| Prerequisites: | Basic knowledge of cloning and SDS-PAGE and immunoblotting; In case of radiolabelling the proteins with 35S-Methionine you must have permission for working with radioisotopes | |
| Teaching goals: |
Learn how to produce your own membrane proteins of interest | |
| Instructor: | Prof. Chris Meisinger and team | |
| Duration: | 2-3 days | |
| Participants: | max. 3 students per course unit | |
| Location: | Institute of Biochemistry and Molecular Biology |
CRISPR-/CAS techniques
Gene knockout using the CRSIPR/CAS system is a powerful technique to silence genes in a quick, reliable and straight-forward fashion. This technique is applicable to various organisms ranging from mammalian cells to bacteria.
| Prerequisites: | Basic knowledge of CRISPR and gene knockout techniques in general, as well as cellular repair mechanisms. | |
| Teaching goals: |
Understanding principles and applications CRISPR-/CAS techniques. | |
| Instructor: | Dr. Claudia Jessen-Trefzer | |
| Duration: | 2 days | |
| Participants: | 3-4 students | |
| Location: | Institute of Pharmaceutical Science (Seminar Room and S2 Laboratory Jessen-Trefzer) |
Electrophysiology of ion channels and transporters
This theory & hands-on course will focus on protein-mediated ion transport across biological membranes. Particular attention will be given to protein reconstitution strategies and the use of Planar Lipid Bilayer versus Solid-supported membrane electrophysiology techniques. Ion transport will be monitored and selectivity, affinity and pKa profiles will be estimated. Students can bring their own samples whenever appropriate.
| Instructor: | Susana Andrade | ||
| Duration: |
3 days | ||
| Participants: | 4 students | ||
| Location: | Institute for Biochemistry (AG Andrade) |
The course runs once during the winter semester, at a time to be agreed by the parts involved and once a minimum number of students raise interest in participating.
EPR-spectroscopy
Electron paramagnetic resonance (EPR) spectroscopy is a most useful and important method to study molecular structures and dynamics. Application to biological molecules provides information on chemical and structural changes in course of a reaction.
|
The course includes:
|
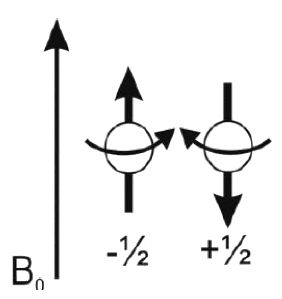 |
|
Prerequisites: |
|
Knowledge of chemical bonds and electron orbitals is helpful, however, knowledge of quantum chemistry is not a prerequisite. | |
|
Teaching goals: |
|
Understanding basic principles of EPR spectroscopy, identification as tool for own research | |
| Instructor: | Prof. Dr. Thorsten Friedrich | ||
| Duration: | 3 days | ||
| Participants: | 6-8 | ||
| Location: | Institute of Biochemistry |

The course is carried out upon request, contact your coordinator.
HPLC/MS and GC/MS methods
HPLC is a powerful method to analyze natural compounds. Modern mashines are often connected to UV spectrometer and mass spectrometer.
| Prerequisites: | Basic knowledge about chemistry | |
| Teaching goals: |
Understanding principles and applications for HPLC analytic | |
| Instructor: | Dr. Roman Makitrynskyy | |
| Duration: | 1 day | |
| Participants: | 3-4 students | |
| Location: | Institute for Pharmaceutical Biology (AG Bechthold) |
In vivo and in vitro localisation of transport proteins
Transport proteins will be stained by fluorescent antibodies. Uptake kinetics and intracellular trafficking will be investigated by confocal microscopy.
| Prerequisites: | Basic knowledge about cell biology | |
| Teaching goals: |
Understanding principles and applications of immunohistochemistry and confocal microscopy | |
| Instructor: | Prof. Dr. Winfried Römer | |
| Duration: | 2 days | |
| Participants: | 2 students | |
| Location: | BIOSS Signalhaus (AG Römer) |
In vivo site directed cross-linking methods
Protein-protein interactions drive many cellular processes, but are often only very transient and difficult to identify in vivo. In vivo cross-linking is a powerful method for stabilizing transient and weak interactions in the context of living cells and is also suitable for down-stream processing of samples by mass spectrometry.
| Prerequisites: | Basic knowledge in genetic engineering and protein biochemistry | |
| Teaching goals: |
Getting an overview over in vivo cross-linking methods and selecting appropriate methods for particular scientific questions | |
| Instructor: | Andreea Andrei, Dr. Yavuz Özturk, Prof. Dr. Hans-Georg Koch | |
| Duration: | 3 days | |
| Participants: | 3-4 students | |
| Location: | Institute for Biochemistry and Molecular Biology (AG Koch) |
Isothermal titration calorimetry to quantify membrane binding of small amphiphilic molecules
ITC has become the gold standard for label-free binding analyses of ligands to receptors. It can also be used for measuring the partitioning of amphiphilic molecules into lipid bilayers, but this requires a special setup and fit model.
| Prerequisites: | Basic knowledge of and interest in biophysical chemistry | |
| Teaching goals: |
Getting to know which problems can be solved by ITC and getting connected to the ITC team | |
| Instructor: | Prof. Dr. Heiko Heerklotz | |
| Duration: | 3 hours incl. lab demonstration | |
| Participants: | max. 5 | |
| Location: |
Institute of Pharmaceutical Sciences – Dept. of Pharmaceutics |
Mass spectrometry on transport proteins
Mass spectrometry (MS) is a key technology in proteomic research which enables the systematic identification and quantification of proteins from cells and tissues. Furthermore, advanced MS-based approaches allow for the characterization of post-translational modifications, biomolecular interactions, structures and functions of proteins. In this course, we will introduce modern proteomic workflows as well as explain the operation principles of different MS instrument types and how to apply them for specific research questions. Hands-on exercises on MS data analysis, visualization and interpretation will help to understand the applications, with a focus on membrane proteins and multi-protein complexes.
| Prerequisites: | Basic knowledge of protein biochemistry. | |
| Teaching goals: |
Understanding MS-based proteomic analyses and applications as well as being able to critically judge the results. | |
| Instructor: | Prof. Dr. Bettina Warscheid, Dr. Friedel Drepper | |
| Duration: | 2-3 days | |
| Participants: | 3-4 students | |
| Location: | Institute of Biology II (Warscheid lab) |
MD-based thermodynamic calculations
The course introduces classical approaches to biomolecules, such as Monte Carlo and molecular dynamics simulations. We discuss force fields and simulation packages, and methods to compute changes in free energies, such as umbrella sampling and thermodynamic integration.
|
Prerequisites: |
Good working knowledge in physical chemistry, basic Linux operating system skills |
|
Teaching goals: |
To perform thermodynamic integration calculations, and to access their quality |
|
Instructor: |
Prof. Thorsten Koslowski |
|
Duration: |
4 x 2 hours |
|
Participants: |
3-4 students |
|
Location: |
Institute for Physical Chemistry (AK Koslowski) |
Membrane dynamics on Giant Liposomes visualized by Confocal microscopy
Synthetic membrane systems, like Giant Liposomes are powerful tools to rebuild cellular processes. Confocal microscopy allows the visualization of receptor protein clustering and membrane invaginations.
| Prerequisites: | Basic knowledge about fluorescence | |
| Teaching goals: |
Understanding principles and applications of Giant Liposomes and Confocal microscopy | |
| Instructor: | Prof. Dr. Winfried Römer | |
| Duration: | 2 days | |
| Participants: | 2 students | |
| Location: | BIOSS Signalhaus (AG Römer) |
Membrane dynamics on supported lipid bilayers visualized by TIRF microscopy
Synthetic membrane systems, like supported lipid bilayers, are powerful tools to rebuild cellular processes. Total internal reflection fluorescence (TIRF) microscopy allows the visualization of protein and lipid clustering, and membrane reorganization in high resolution
| Prerequisites: | Basic knowledge about fluorescence | |
| Teaching goals: |
Understanding principles and applications of supported lipid bilayers and TIRF microscopy | |
| Instructor: | Prof. Dr. Winfried Römer | |
| Duration: | 2 days | |
| Participants: | 2 students | |
| Location: | BIOSS Signalhaus (AG Römer) |
Microcalorimetry
Microcalorimetric techniques monitor very small heat responses of a sample after injection of a second component (ITC), during a temperature scan (DSC), pressure change (PPC), or due to intrinsic (e.g., degradation) processes (isothermal calorimetry). The course gives an overview of principles, instruments and applications followed by a short practical training in the lab.
| Prerequisites: | Basic knowledge of and interest in biophysical chemistry | |
| Teaching goals: |
Understanding what microcalorimetry can do for your project | |
| Instructor: | Prof. Dr. Heiko Heerklotz | |
| Duration: | 3 hours (2 hours seminar + 1 h practical training at a Malvern VP DSC) | |
| Participants: | max. 5 | |
| Location: | Institute of Pharmaceutical Sciences – Dept. of Pharmaceutics |
Molecular modeling
The molecular modeling course introduces into computational methods for modeling biomolecules and their interactions. This includes the modeling of protein structures (homology modeling), the modeling of direct interactions (protein and ligand docking) as well as the simulation of the molecular dynamics of proteins, e.g. for modeling of conformational changes of proteins.
| Prerequisites: | - | |
| Teaching goals: |
Skills should be learned that enable the application of computational modelling techniques to support the individual research projects | |
| Instructor: | Jianyu Li, Aurelien Moumbock, Prof. Dr. Stefan Günther | |
| Duration: | 1,5 days | |
| Participants: | 25 (maximal number) | |
| Location: | Rechenzentrum, Hermann-Herder Straße 9, Uni-Freiburg |
Peripheral Peptides and Proteins at Lipid Monolayers
Lipid monolayers are models for membrane surfaces, where the lateral pressure can be controlled. Adsorption of proteins can expand or compress the monolayer depending on the local structure and interaction and on the pre-set lateral pressure.
| Prerequisites: | Basic knowledge of and interest in biophysical chemistry | |
| Teaching goals: |
Getting to know about possible applications of Langmuir film balances | |
| Instructor: | Dr. Maria Hoernke | |
| Duration: | 3 hours | |
| Participants: | max. 5 | |
| Location: | Institute of Pharmaceutical Sciences – Dept. of Pharmaceutics |
Protein Structure Determination by X-ray Crystallography
Crystal structures remain the predominant way of investigating and visualizing molecular features in atomic detail. In this method course we have a guided walk-through of a complete structure solution process from a diffraction image to a refined structural model.
| Prerequisites: | Basic understanding of 3D structures and molecular interactions. Ideally: personal laptop for installation of (freely available) software. | |
| Teaching goals: |
Basic understanding of the steps involved in the processing of diffraction images, the preparation of a data set and the solution of a three-dimensional structure by molecular replacement. | |
| Instructor: | Dr. Stefan Gerhardt | |
| Duration: | 4-5 days |
|
| Participants: | 4-6 students | |
| Location: | Institute for Biochemistry (AG Einsle) | |
Synthetic Biology of Membrane Proteins
Synthetic biology describes the design and engineering of proteins/enzymes with novel properties that are usually not found in nature. It is a very powerful approach to generate novel proteins, delineate evolutionary principles and for understanding the role of protein-protein interactions in biological systems.
The course includes theoretical and experimental parts:
Theoretical Part:
- Background on synthetic biology with special emphasis on membrane proteins
- Recent advances and applications of synthetic membrane proteins.
Experimental Part:
- Experimental design and cloning of genetically fused and truncated version of membrane proteins. As examples a fusion between the P1B-ATPase type copper transporter CcoI and the periplasmic copper chaperon SenC will be generated. In addition, we will use a reductive approach of synthetic biology by constructing a truncated version of the recently identified cupric reductase CcoG, lacking the periplasmic immunoglobulin domain.
- The functionality of these constructs will be phenotypically analyzed in vivo and in vitro.
Teaching goals: Understanding basic principles for designing synthetic membrane proteins.
Instructor: Dr. Yavuz Öztürk
Duration: 3 days
Participants: 3-4 students
Location: Institute of Biochemistry and Molecular Biology
Time resolved fluorescence anisotropy of membrane probes to monitor membrane order and dynamics
The course gives a short introduction to the principles of fluorescence spectroscopy and its applications in membrane biophysics, covering e.g. steady-state fluorescence spectroscopy, time-resolved fluorescence spectroscopy, anisotropy, quenching, and Förster resonance energy transfer (FRET). Special focus is on time-correlated single photon counting and its advantages over steady-state measurements.
| Prerequisites: | Basic knowledge of and interest in biophysical chemistry | |
| Teaching goals: |
Becoming aware of the opportunities of fluorescence, particularly TCSPC | |
| Instructor: | Prof. Dr. Heiko Heerklotz, Johannes Schnur | |
| Duration: | 3 hours (2 hours seminar + 1 h practical training at a Malvern VP DSC) | |
| Participants: | max. 5 | |
| Location: | Institute of Pharmaceutical Sciences – Dept. of Pharmaceutics |



