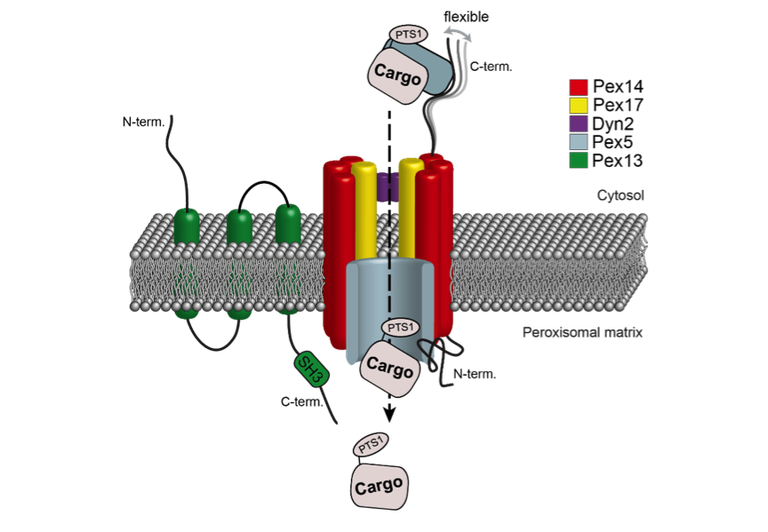
Transport processes across peroxisomal membranes
Peroxisomes are highly dynamic metabolic organelles that fulfill various essential physiological functions in nucleated eukaryotic cells. Their importance for cellular fitness is manifested in the occurrence of severe, often lethal diseases in humans lacking functional peroxisomes. Biogenesis and functions of peroxisomes rely on the import of nucleus-encoded proteins from the cytosol. Proteins of the peroxisomal matrix are generally recognized in the cytosol by specific receptors via an N-terminal peroxisomal targeting signal (PTS) 1 or a PTS2 at the C-terminus [1]. At the peroxisomal membrane, cargo-loaded receptor proteins bind to a docking complex consisting of Pex14, Pex13 and, in the yeast Saccharomyces cerevisiae, Pex17 and Dyn2 [2, 3]. By an integrative structural approach combining native MS, chemical crosslinking and cryo-electron microscopy we showed that the reconstituted Pex14/Pex17 complex resembles a rod-like structure with a Pex14 homotrimer and a single Pex17 monomer in a parallel arrangement [4] (Figure 1). Following docking, the cargo protein is translocated across the peroxisomal membrane into the matrix through PTS1- and PTS2-specific import pores, both containing Pex14 and either the PTS1 receptor Pex5 or the PTS2 co-receptor Pex18, and Pex17 [1].
Figure 1. Schematic model of the transport of newly synthesized peroxisomal matrix proteins comprising a peroxisomal targeting sequence 1 (PTS1) across the peroxisomal membrane. Pex14 and Pex17 assemble into rod-like particles with a 3:1 stoichiometry that are involved in the docking of receptor-cargo complexes to the peroxisomal membrane followed by cargo import through a transiently formed PTS1 import pore.
We have proposed that phosphorylation plays an important role in the modulation of peroxisome dynamics [5]. On the basis of phosphosite mapping [6], we discovered that import of peroxisomal matrix proteins is modulated by phosphorylation of central elements of the import pores. Phosphorylation of Pex5 in the PTS1-binding region reduces the binding affinity to specific cargo proteins (data unpublished) and site-specific phosphorylation of Pex14 specifically affects import of peroxisomal citrate synthase 2 in a carbon source-dependent manner [7]. Thus, our findings highlight the importance of peroxin phosphorylation for the regulation of protein transport processes into peroxisomes.
In addition to protein import, peroxisome biogenesis and proliferation require the transfer of lipids from the ER. We identified Pex30 in S. cerevisiae to be involved in the formation of ER-peroxisome contact sites (EPCONs) [8]. In mammals, the peroxisomal membrane protein acyl-CoA-binding domain protein 5, ACBD5, establishes EPCONs by binding to vesicle-associated membrane protein-associated proteins (VAP) A and B located in the ER membrane [9]. We identified Scs2, a yeast homolog of VAP proteins that is known to be involved in intracellular phospholipid lipid transport [10], to be associated with specific peroxisomal membrane proteins, suggesting that they together establish EPCONs in S. cerevisiae.
The focus of this research project lies on structural studies of Pex14-centered protein complexes for an improved molecular understanding of protein transport processes into peroxisomes and on the study of formation and regulation of EPCONs with a potential role in lipid transfer to peroxisomes.
References
[1] Walter T, Erdmann R (2019) Protein J. 38, 351-362.
[2] Oeljeklaus S, Reinartz B, Wolf J, Wiese S, Tonillo J, Podwojski K, Kuhlmann K, Stephan C, Meyer HE, Schliebs W, Brocard C, Erdmann R, Warscheid B (2012) J. Proteome Res. 11, 2567-2580.
[3] Chan A, Schummer A, Fischer S, Bender J, Drepper F, Oeljeklaus S, Kunau WH, Girzalsky W, Warscheid B*, Erdmann R* (2016) Eur. J. Cell Biol. 95, 585-597. *corresponding
[4] Lill P*, Hansen T*, Wendscheck D*, Klink BU, Jeziorek T, Miehling J, Bender J, Drepper F, Girzalsky W, Warscheid B#, Erdmann R#, Gatsogiannis C (2020) bioRxiv 854497 (doi.org/10.1101/854497). *equal first authorship, #equal contribution
[5] Oeljeklaus S, Schummer A, Mastalski T, Platta HW, Warscheid B (2016) Biochim. Biophys. Acta 1863, 1027-1037
[6] Schummer A, Fischer S, Oeljeklaus S, Warscheid B (2017) Methods Mol. Biol. 1595, 267-289.
[7] Schummer A*, Maier R*, Gabay-Maskit S, Hansen T, Muehlhaeuser WWD, Suppanz I, Fadel A, Schuldiner M, Girzalsky W, Oeljeklaus S, Zalckvar E, Erdmann R, Warscheid B (2020) bioRxiv 186833 (doi.org/10.1101/2020.07.03.186833). *equal first authorship
[8] David C, Koch JP, Oeljeklaus S, Laernsack A, Melchior S, Wiese S, Schummer A, Erdmann R, Warscheid B*, Brocard C* (2013) Mol. Cell. Proteomics 12, 2408-2425. *corresponding
[9] Costello JL, Castro IG, Hacker C, Schrader TA, Metz J, Zeuschner D, Azadi AS, Godinho LF, Costina V, Findeisen P, Manner A, Islinger M, Schrader M. (2017) J. Cell Biol. 216, 331-342.
[10] Loewen CJR, Roy A, Levine, TP (2003) EMBO J. 22, 2025-2035.
Contact
Prof. Dr. Bettina Warscheid
Institute of Biology II
Schänzlestr. 1
79104 Freiburg
Phone: +49 (761) 203 2690
Fax: +49 (761) 203 2601
Bettina.Warscheid@biologie.uni-freiburg.de
https://www.proteomics.uni-freiburg.de/team/Prof.%20Dr.%20Bettina%20Warscheid
http://www.bioss.uni-freiburg.de/prof-bettina-warscheid/
https://www.cibss.uni-freiburg.de/people/principle-investigators/prof-dr-bettina-warscheid/