Elucidation of the mechanism and biological role of selected GPCRs
G-protein coupled receptors (GPCRs) are the largest superfamily of transmembrane proteins that transport information across the membrane. They are able to bind an extraordinary variety of ligands, ranging from small molecules to large proteins, and pass this binding event across the cell membrane to a much smaller group of signaling proteins inside the cell. The class of GPCR-proteins contains several famous drug targets. Binding site similarities between the homologue proteins have an important effect on drug responses because single drugs may elicit an activation or inhibition of several GPCRs in parallel (Fig.1) [1]. However, the majority (56%) of non-olfactory GPCRs have a broad untapped therapeutic potential, particularly in genetic and immune system disorders.
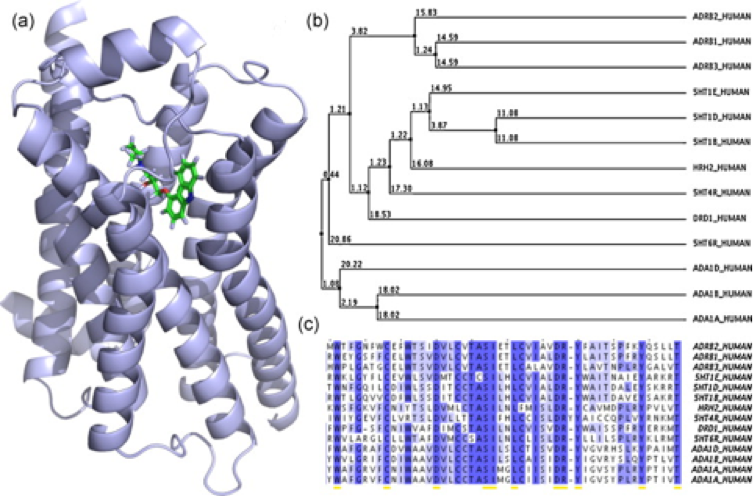
Figure 1: (a) Crystal structure of the beta-2-adrenergic receptor (ADRB2) with its inverse agonist carazolol (PDB-ID: 2rhi). (b) Average distance tree of 13 G protein-coupled receptors (GPCRs) represented by entry names of the UniProt database. (c) ClustalW multiple sequence alignment of a part of the receptors ligand/drug binding sites [1].
The allosteric behavior of most GPCRs (conformational changes of the binding sites as well as of the global receptors) will be analyzed by Molecular Dynamics (MD) simulations. A series of large scale simulations followed by a conformational landscape mapping allows for the mechanistic understanding of the structural dynamics of the interplay between ligand binding and induction of conformational changes. The Günther group has shown, e.g. for the tyrosine kinase Lck and for the kinase JNK3, that modelling of biomolecular interactions and long time simulations can point out the mechanism of action and global conformational changes that reveal also novel allosteric binding sites for drugs [2, 3].
For many GPCRs the structure has not been solved yet and the clarification of the mechanism of action as well as the development of novel binders is an ambitious and promising challenge. The elucidation of the mechanism of action and the overall design process of chemical probes will mainly be driven by structural modelling of the receptors and their interactions.
Within this project chemical probes will be developed to modulate GPCR signaling and to analyze their biological role and therapeutic potential. Novel binders will be designed that can be further developed into Proteolysis Targeting Chimeras (PROTACs).
Modelling of transmembrane transport of small molecules
The P-glycoprotein (P-gp) is capable of effluxing a broad range of cytosolic and membrane penetrating xenobiotic substrates, thus leading to multidrug resistance and posing a threat for the therapeutic treatment of several diseases, including cancer and central nervous disorders. In a preliminary work, a virtual screening campaign followed by experimental validation in Caco-2, MDKCII, and MDKCII mdr1 transfected cell lines has been conducted for the identification of novel phospholipids with P-gp transportation inhibitory activity. C8:0 Phosphatidylinositol-sodium salt (PI) was found to significantly inhibit transmembrane P-gp transportation in vitro in a reproducible-, cell line-, and substrate-independent manner [4]. With additional modeling efforts it will be determined whether also other compound classes come into question for the co-administration with oral drugs to successfully increase their bioavailability.
References
[1] R. Adams, C.L. Worth, S. Günther, M. Dunkel, R. Lehmann, R. Preissner. Binding sites in membrane proteins--diversity, druggability and prospects. Eur J Cell Biol. 2012 Apr;91(4):326-39.
[2] F. A. Hartl, E. Beck-Garcìa, N. M. Woessner, L. J. Flachsmann, R. M.-H. Velasco Cárdenas, S. M. Brandl , S. Taromi, G. J. Fiala, A. Morath, P. Mishra, O. S. Yousefi, J. Zimmermann, N. Hoefflin, M. Köhn , B. Wöhrl, R. Zeiser, K. Schweimer, S. Günther, W. W. Schamel , and S. Minguet. Noncanonical binding of Lck to CD3ε promotes TCR signaling and CAR function. Nat. Immunol. (2020, in press)
[3] P. Mishra, S. Günther. New insights into the structural dynamics of the kinase JNK3 Sci Rep. 2018; 8: 9435.
[4] X. Lucas, S. Simon, R. Schubert, S. Günther. Discovery of the inhibitory effect of a phosphatidylinositol derivative on P-glycoprotein by virtual screening followed by in vitro cellular studies. PLoS One. 2013 Apr 9;8(4):e60679.
Contact
Prof. Dr. Stefan Günther
Pharmaceutical Bioinformatics
Hermann-Herder-Str. 9
79104 Freiburg
Phone: +49 (761) 203 4871
Fax: +49 (761) 203 97769
stefan.guenther@pharmazie.uni-freiburg.de



